Practical session on xarray¶
Instructions¶
Load the
python-training/misc/data/mesh_mask_eORCA1_v2.2.ncfile usingopen_dataset.
Extract the surface (
z=0) and first time step (t=0) usingisel
Plot the land-sea mask (
tmask) variable.
Compute the cell surface (
e1t x e2t)
Load the
python-training/misc/data/surface_thato.ncfile usingopen_dataset.
Extract the SST (
thetaovariable) at the surface (olevel=0)
Compute and display the time-average SST
Compute the mean SST over years 1958-1962
Compute the mean over years 2014-2018
Plot the SST difference between 2018-2014 and 1958-1962
Compute the SST global mean time-series (weight by cell surface $e1t \times e2t$)
Remove the monthly clim from the time-series using
groupyontime_counter.month
Compute the rolling mean of the time-series, using a 3-year window. Plot the raw and smoothed anomalies
Corrections¶
Load the
python-training/misc/data/mesh_mask_eORCA1_v2.2.ncfile usingopen_dataset.
import xarray as xr
mesh = xr.open_dataset('data/mesh_mask_eORCA1_v2.2.nc')
mesh
<xarray.Dataset>
Dimensions: (t: 1, y: 332, x: 362, z: 75)
Dimensions without coordinates: t, y, x, z
Data variables:
e1t (t, y, x) float64 ...
e2t (t, y, x) float64 ...
e3t_0 (t, z, y, x) float64 ...
glamf (t, y, x) float32 ...
glamt (t, y, x) float32 ...
gphif (t, y, x) float32 ...
gphit (t, y, x) float32 ...
tmask (t, z, y, x) int8 ...
Attributes:
file_name: mesh_mask.nc
TimeStamp: 08/11/2019 15:49:38 +0100
history: Tue Aug 24 09:30:40 2021: ncks -O -v tmask,glamf,gphif,gphit,...
NCO: netCDF Operators version 4.9.1 (Homepage = http://nco.sf.net,...- t: 1
- y: 332
- x: 362
- z: 75
- e1t(t, y, x)float64...
[120184 values with dtype=float64]
- e2t(t, y, x)float64...
[120184 values with dtype=float64]
- e3t_0(t, z, y, x)float64...
[9013800 values with dtype=float64]
- glamf(t, y, x)float32...
[120184 values with dtype=float32]
- glamt(t, y, x)float32...
[120184 values with dtype=float32]
- gphif(t, y, x)float32...
[120184 values with dtype=float32]
- gphit(t, y, x)float32...
[120184 values with dtype=float32]
- tmask(t, z, y, x)int8...
[9013800 values with dtype=int8]
- file_name :
- mesh_mask.nc
- TimeStamp :
- 08/11/2019 15:49:38 +0100
- history :
- Tue Aug 24 09:30:40 2021: ncks -O -v tmask,glamf,gphif,gphit,glamt,e1t,e2t,e3t_0 save_mesh_mask_eORCA1_v2.2.nc -L 9 mesh_mask_eORCA1_v2.2.nc Tue Aug 24 09:29:35 2021: ncks -4 save_mesh_mask_eORCA1_v2.2.nc save_mesh_mask_eORCA1_v2.2.nc
- NCO :
- netCDF Operators version 4.9.1 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)
Extract the surface (
z=0) and first time step (t=0) usingisel
mesh = mesh.isel(z=0, t=0)
mesh
<xarray.Dataset>
Dimensions: (y: 332, x: 362)
Dimensions without coordinates: y, x
Data variables:
e1t (y, x) float64 ...
e2t (y, x) float64 ...
e3t_0 (y, x) float64 ...
glamf (y, x) float32 ...
glamt (y, x) float32 ...
gphif (y, x) float32 ...
gphit (y, x) float32 ...
tmask (y, x) int8 ...
Attributes:
file_name: mesh_mask.nc
TimeStamp: 08/11/2019 15:49:38 +0100
history: Tue Aug 24 09:30:40 2021: ncks -O -v tmask,glamf,gphif,gphit,...
NCO: netCDF Operators version 4.9.1 (Homepage = http://nco.sf.net,...- y: 332
- x: 362
- e1t(y, x)float64...
[120184 values with dtype=float64]
- e2t(y, x)float64...
[120184 values with dtype=float64]
- e3t_0(y, x)float64...
[120184 values with dtype=float64]
- glamf(y, x)float32...
[120184 values with dtype=float32]
- glamt(y, x)float32...
[120184 values with dtype=float32]
- gphif(y, x)float32...
[120184 values with dtype=float32]
- gphit(y, x)float32...
[120184 values with dtype=float32]
- tmask(y, x)int8...
[120184 values with dtype=int8]
- file_name :
- mesh_mask.nc
- TimeStamp :
- 08/11/2019 15:49:38 +0100
- history :
- Tue Aug 24 09:30:40 2021: ncks -O -v tmask,glamf,gphif,gphit,glamt,e1t,e2t,e3t_0 save_mesh_mask_eORCA1_v2.2.nc -L 9 mesh_mask_eORCA1_v2.2.nc Tue Aug 24 09:29:35 2021: ncks -4 save_mesh_mask_eORCA1_v2.2.nc save_mesh_mask_eORCA1_v2.2.nc
- NCO :
- netCDF Operators version 4.9.1 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)
Plot the land-sea mask (
tmask) variable.
tmask = mesh['tmask']
tmask.plot()
<matplotlib.collections.QuadMesh at 0x7f3ce4b62e90>
Compute the cell surface (
e1t x e2t)
surface = mesh['e1t'] * mesh['e2t']
surface
<xarray.DataArray (y: 332, x: 362)>
array([[1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ...,
1.60000000e+01, 1.60000000e+01, 1.60000000e+01],
[1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ...,
1.60000000e+01, 1.60000000e+01, 1.60000000e+01],
[1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ...,
1.60000000e+01, 1.60000000e+01, 1.60000000e+01],
...,
[4.96784478e+07, 4.96784478e+07, 1.68195300e+08, ...,
1.68195300e+08, 4.96784478e+07, 4.96784478e+07],
[3.18035791e+07, 3.18035791e+07, 1.55003643e+08, ...,
1.55003643e+08, 3.18035791e+07, 3.18035791e+07],
[3.18035791e+07, 3.18035791e+07, 1.55003643e+08, ...,
1.55003643e+08, 3.18035791e+07, 3.18035791e+07]])
Dimensions without coordinates: y, x- y: 332
- x: 362
- 16.0 16.0 16.0 16.0 16.0 ... 2.558e+08 1.55e+08 3.18e+07 3.18e+07
array([[1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ..., 1.60000000e+01, 1.60000000e+01, 1.60000000e+01], [1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ..., 1.60000000e+01, 1.60000000e+01, 1.60000000e+01], [1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ..., 1.60000000e+01, 1.60000000e+01, 1.60000000e+01], ..., [4.96784478e+07, 4.96784478e+07, 1.68195300e+08, ..., 1.68195300e+08, 4.96784478e+07, 4.96784478e+07], [3.18035791e+07, 3.18035791e+07, 1.55003643e+08, ..., 1.55003643e+08, 3.18035791e+07, 3.18035791e+07], [3.18035791e+07, 3.18035791e+07, 1.55003643e+08, ..., 1.55003643e+08, 3.18035791e+07, 3.18035791e+07]])
Here the output DatarArray has no name. You can give him one as follows:
surface.name = 'surface'
surface
<xarray.DataArray 'surface' (y: 332, x: 362)>
array([[1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ...,
1.60000000e+01, 1.60000000e+01, 1.60000000e+01],
[1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ...,
1.60000000e+01, 1.60000000e+01, 1.60000000e+01],
[1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ...,
1.60000000e+01, 1.60000000e+01, 1.60000000e+01],
...,
[4.96784478e+07, 4.96784478e+07, 1.68195300e+08, ...,
1.68195300e+08, 4.96784478e+07, 4.96784478e+07],
[3.18035791e+07, 3.18035791e+07, 1.55003643e+08, ...,
1.55003643e+08, 3.18035791e+07, 3.18035791e+07],
[3.18035791e+07, 3.18035791e+07, 1.55003643e+08, ...,
1.55003643e+08, 3.18035791e+07, 3.18035791e+07]])
Dimensions without coordinates: y, x- y: 332
- x: 362
- 16.0 16.0 16.0 16.0 16.0 ... 2.558e+08 1.55e+08 3.18e+07 3.18e+07
array([[1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ..., 1.60000000e+01, 1.60000000e+01, 1.60000000e+01], [1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ..., 1.60000000e+01, 1.60000000e+01, 1.60000000e+01], [1.60000000e+01, 1.60000000e+01, 1.60000000e+01, ..., 1.60000000e+01, 1.60000000e+01, 1.60000000e+01], ..., [4.96784478e+07, 4.96784478e+07, 1.68195300e+08, ..., 1.68195300e+08, 4.96784478e+07, 4.96784478e+07], [3.18035791e+07, 3.18035791e+07, 1.55003643e+08, ..., 1.55003643e+08, 3.18035791e+07, 3.18035791e+07], [3.18035791e+07, 3.18035791e+07, 1.55003643e+08, ..., 1.55003643e+08, 3.18035791e+07, 3.18035791e+07]])
Load the
python-training/misc/data/surface_thato.ncfile usingopen_dataset.
data = xr.open_dataset('data/surface_thetao.nc')
data
<xarray.Dataset>
Dimensions: (y: 332, x: 362, nvertex: 4, olevel: 1, axis_nbounds: 2, time_counter: 732)
Coordinates:
nav_lat (y, x) float32 ...
nav_lon (y, x) float32 ...
* olevel (olevel) float32 0.5058
time_centered (time_counter) object 1958-01-16 12:00:00 ... 2018-...
* time_counter (time_counter) object 1958-01-16 12:00:00 ... 2018-...
Dimensions without coordinates: y, x, nvertex, axis_nbounds
Data variables:
bounds_nav_lat (y, x, nvertex) float32 ...
bounds_nav_lon (y, x, nvertex) float32 ...
olevel_bounds (olevel, axis_nbounds) float32 0.0 1.024
thetao (time_counter, olevel, y, x) float32 ...
time_centered_bounds (time_counter, axis_nbounds) object 1958-01-01 00:0...
time_counter_bounds (time_counter, axis_nbounds) object 1958-01-01 00:0...
Attributes:
name: GCB-eORCA1-JRA14-CO2ANTH_1m_grid_T
description: Created by xios
title: Created by xios
Conventions: CF-1.6
timeStamp: 2019-Oct-25 14:55:39 GMT
uuid: dfe6b939-ae41-42de-977a-3a151c062065
LongName: ORCA1_LIM3_PISCES NEMO configuration
history: Tue Aug 24 09:26:54 2021: ncks -L 9 surface_th...
NCO: netCDF Operators version 4.9.1 (Homepage = htt...
nco_openmp_thread_number: 1- y: 332
- x: 362
- nvertex: 4
- olevel: 1
- axis_nbounds: 2
- time_counter: 732
- nav_lat(y, x)float32...
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- bounds :
- bounds_nav_lat
[120184 values with dtype=float32]
- nav_lon(y, x)float32...
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- bounds :
- bounds_nav_lon
[120184 values with dtype=float32]
- olevel(olevel)float320.5058
- name :
- olevel
- long_name :
- Vertical T levels
- units :
- m
- positive :
- down
- bounds :
- olevel_bounds
array([0.50576], dtype=float32)
- time_centered(time_counter)object...
- standard_name :
- time
- long_name :
- Time axis
- time_origin :
- 1958-01-01 00:00:00
- bounds :
- time_centered_bounds
array([cftime.DatetimeNoLeap(1958, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 3, 16, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(2018, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 12, 16, 12, 0, 0, 0, has_year_zero=True)], dtype=object) - time_counter(time_counter)object1958-01-16 12:00:00 ... 2018-12-...
- axis :
- T
- standard_name :
- time
- long_name :
- Time axis
- time_origin :
- 1958-01-01 00:00:00
- bounds :
- time_counter_bounds
array([cftime.DatetimeNoLeap(1958, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 3, 16, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(2018, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 12, 16, 12, 0, 0, 0, has_year_zero=True)], dtype=object)
- bounds_nav_lat(y, x, nvertex)float32...
[480736 values with dtype=float32]
- bounds_nav_lon(y, x, nvertex)float32...
[480736 values with dtype=float32]
- olevel_bounds(olevel, axis_nbounds)float32...
- units :
- m
array([[0. , 1.023907]], dtype=float32)
- thetao(time_counter, olevel, y, x)float32...
- standard_name :
- sea_water_conservative_temperature
- long_name :
- sea_water_potential_temperature
- units :
- degree_C
- online_operation :
- average
- interval_operation :
- 1 month
- interval_write :
- 1 month
- cell_methods :
- time: mean
- cell_measures :
- area: area
[87974688 values with dtype=float32]
- time_centered_bounds(time_counter, axis_nbounds)object...
array([[cftime.DatetimeNoLeap(1958, 1, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 2, 1, 0, 0, 0, 0, has_year_zero=True)], [cftime.DatetimeNoLeap(1958, 2, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 3, 1, 0, 0, 0, 0, has_year_zero=True)], [cftime.DatetimeNoLeap(1958, 3, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 4, 1, 0, 0, 0, 0, has_year_zero=True)], ..., [cftime.DatetimeNoLeap(2018, 10, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 11, 1, 0, 0, 0, 0, has_year_zero=True)], [cftime.DatetimeNoLeap(2018, 11, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 12, 1, 0, 0, 0, 0, has_year_zero=True)], [cftime.DatetimeNoLeap(2018, 12, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2019, 1, 1, 0, 0, 0, 0, has_year_zero=True)]], dtype=object) - time_counter_bounds(time_counter, axis_nbounds)object...
array([[cftime.DatetimeNoLeap(1958, 1, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 2, 1, 0, 0, 0, 0, has_year_zero=True)], [cftime.DatetimeNoLeap(1958, 2, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 3, 1, 0, 0, 0, 0, has_year_zero=True)], [cftime.DatetimeNoLeap(1958, 3, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 4, 1, 0, 0, 0, 0, has_year_zero=True)], ..., [cftime.DatetimeNoLeap(2018, 10, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 11, 1, 0, 0, 0, 0, has_year_zero=True)], [cftime.DatetimeNoLeap(2018, 11, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 12, 1, 0, 0, 0, 0, has_year_zero=True)], [cftime.DatetimeNoLeap(2018, 12, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2019, 1, 1, 0, 0, 0, 0, has_year_zero=True)]], dtype=object)
- name :
- GCB-eORCA1-JRA14-CO2ANTH_1m_grid_T
- description :
- Created by xios
- title :
- Created by xios
- Conventions :
- CF-1.6
- timeStamp :
- 2019-Oct-25 14:55:39 GMT
- uuid :
- dfe6b939-ae41-42de-977a-3a151c062065
- LongName :
- ORCA1_LIM3_PISCES NEMO configuration
- history :
- Tue Aug 24 09:26:54 2021: ncks -L 9 surface_thetao.nc surface_thetao.nc Tue Aug 24 09:26:21 2021: ncrcat surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19580101_19581231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19590101_19591231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19600101_19601231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19610101_19611231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19620101_19621231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19630101_19631231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19640101_19641231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19650101_19651231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19660101_19661231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19670101_19671231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19680101_19681231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19690101_19691231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19700101_19701231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19710101_19711231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19720101_19721231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19730101_19731231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19740101_19741231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19750101_19751231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19760101_19761231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19770101_19771231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19780101_19781231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19790101_19791231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19800101_19801231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19810101_19811231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19820101_19821231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19830101_19831231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19840101_19841231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19850101_19851231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19860101_19861231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19870101_19871231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19880101_19881231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19890101_19891231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19900101_19901231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19910101_19911231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19920101_19921231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19930101_19931231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19940101_19941231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19950101_19951231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19960101_19961231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19970101_19971231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19980101_19981231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19990101_19991231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20000101_20001231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20010101_20011231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20020101_20021231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20030101_20031231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20040101_20041231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20050101_20051231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20060101_20061231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20070101_20071231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20080101_20081231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20090101_20091231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20100101_20101231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20110101_20111231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20120101_20121231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20130101_20131231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20140101_20141231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20150101_20151231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20160101_20161231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20170101_20171231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_20180101_20181231_1M_grid_T.nc surface_thetao.nc Tue Jun 29 12:26:28 2021: ncks -d olevel,0 -v thetao nico_GCB-eORCA1-JRA14-CO2ANTH_19580101_19581231_1M_grid_T.nc surface_nico_GCB-eORCA1-JRA14-CO2ANTH_19580101_19581231_1M_grid_T.nc Tue Dec 10 11:48:40 2019: ncks -v e3t,thetao,so /ccc/store/cont003/thredds/p48ethe/ORCA1_LIM3_PISCES/DEVT/ORCA1ia/GCB-eORCA1-JRA14-CO2ANTH/OCE/Output/MO/GCB-eORCA1-JRA14-CO2ANTH_19580101_19581231_1M_grid_T.nc /data/rperson/NICO/nico_GCB-eORCA1-JRA14-CO2ANTH_19580101_19581231_1M_grid_T.nc Fri Oct 25 17:50:36 2019: ncrcat -C --buffer_size 838860800 -p /ccc/scratch/cont003/gen0040/p48ethe/IGCM_OUT/ORCA1_LIM3_PISCES/PROD/ORCA1ia/GCB-eORCA1-JRA14-CO2ANTH/OCE/Output/MO GCB-eORCA1-JRA14-CO2ANTH_19580101_19581231_1M_grid_T.nc --output GCB-eORCA1-JRA14-CO2ANTH_19580101_19581231_1M_grid_T.nc
- NCO :
- netCDF Operators version 4.9.1 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)
- nco_openmp_thread_number :
- 1
Extract the SST (
thetaovariable) at the surface (olevel=0)
thetao = data['thetao'].isel(olevel=0)
thetao
<xarray.DataArray 'thetao' (time_counter: 732, y: 332, x: 362)>
[87974688 values with dtype=float32]
Coordinates:
nav_lat (y, x) float32 ...
nav_lon (y, x) float32 ...
olevel float32 0.5058
time_centered (time_counter) object 1958-01-16 12:00:00 ... 2018-12-16 1...
* time_counter (time_counter) object 1958-01-16 12:00:00 ... 2018-12-16 1...
Dimensions without coordinates: y, x
Attributes:
standard_name: sea_water_conservative_temperature
long_name: sea_water_potential_temperature
units: degree_C
online_operation: average
interval_operation: 1 month
interval_write: 1 month
cell_methods: time: mean
cell_measures: area: area- time_counter: 732
- y: 332
- x: 362
- ...
[87974688 values with dtype=float32]
- nav_lat(y, x)float32...
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- bounds :
- bounds_nav_lat
[120184 values with dtype=float32]
- nav_lon(y, x)float32...
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- bounds :
- bounds_nav_lon
[120184 values with dtype=float32]
- olevel()float320.5058
- name :
- olevel
- long_name :
- Vertical T levels
- units :
- m
- positive :
- down
- bounds :
- olevel_bounds
array(0.50576, dtype=float32)
- time_centered(time_counter)object1958-01-16 12:00:00 ... 2018-12-...
- standard_name :
- time
- long_name :
- Time axis
- time_origin :
- 1958-01-01 00:00:00
- bounds :
- time_centered_bounds
array([cftime.DatetimeNoLeap(1958, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 3, 16, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(2018, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 12, 16, 12, 0, 0, 0, has_year_zero=True)], dtype=object) - time_counter(time_counter)object1958-01-16 12:00:00 ... 2018-12-...
- axis :
- T
- standard_name :
- time
- long_name :
- Time axis
- time_origin :
- 1958-01-01 00:00:00
- bounds :
- time_counter_bounds
array([cftime.DatetimeNoLeap(1958, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 3, 16, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(2018, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 12, 16, 12, 0, 0, 0, has_year_zero=True)], dtype=object)
- standard_name :
- sea_water_conservative_temperature
- long_name :
- sea_water_potential_temperature
- units :
- degree_C
- online_operation :
- average
- interval_operation :
- 1 month
- interval_write :
- 1 month
- cell_methods :
- time: mean
- cell_measures :
- area: area
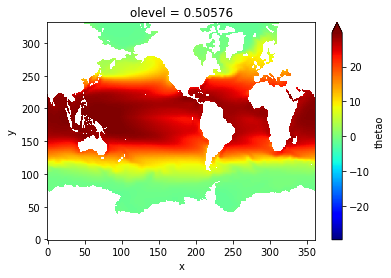
Compute and display the time-average SST
import matplotlib.pyplot as plt
plt.rcParams['text.usetex'] = False
theta_mean = thetao.mean(dim='time_counter')
theta_mean.plot(robust=True, cmap=plt.cm.jet)
<matplotlib.collections.QuadMesh at 0x7f3ce366be90>
Compute the mean SST over years 1958-1962
sst_early = thetao.sel(time_counter=slice('1958-01-01', '1962-12-31')).mean(dim='time_counter')
sst_early
<xarray.DataArray 'thetao' (y: 332, x: 362)>
array([[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]], dtype=float32)
Coordinates:
nav_lat (y, x) float32 ...
nav_lon (y, x) float32 ...
olevel float32 0.5058
Dimensions without coordinates: y, x- y: 332
- x: 362
- nan nan nan nan nan nan nan nan ... nan nan nan nan nan nan nan nan
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - nav_lat(y, x)float32...
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- bounds :
- bounds_nav_lat
[120184 values with dtype=float32]
- nav_lon(y, x)float32...
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- bounds :
- bounds_nav_lon
[120184 values with dtype=float32]
- olevel()float320.5058
- name :
- olevel
- long_name :
- Vertical T levels
- units :
- m
- positive :
- down
- bounds :
- olevel_bounds
array(0.50576, dtype=float32)
Compute the mean over years 2014-2018
sst_late = thetao.sel(time_counter=slice('2014-01-01', '2018-12-31')).mean(dim='time_counter')
sst_late
<xarray.DataArray 'thetao' (y: 332, x: 362)>
array([[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]], dtype=float32)
Coordinates:
nav_lat (y, x) float32 ...
nav_lon (y, x) float32 ...
olevel float32 0.5058
Dimensions without coordinates: y, x- y: 332
- x: 362
- nan nan nan nan nan nan nan nan ... nan nan nan nan nan nan nan nan
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], dtype=float32) - nav_lat(y, x)float32...
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- bounds :
- bounds_nav_lat
[120184 values with dtype=float32]
- nav_lon(y, x)float32...
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- bounds :
- bounds_nav_lon
[120184 values with dtype=float32]
- olevel()float320.5058
- name :
- olevel
- long_name :
- Vertical T levels
- units :
- m
- positive :
- down
- bounds :
- olevel_bounds
array(0.50576, dtype=float32)
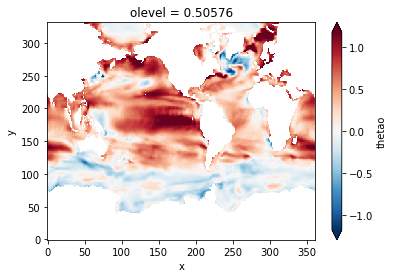
Plot the SST difference between 2014-2018 and 1958-1962
(sst_late - sst_early).plot(robust=True)
<matplotlib.collections.QuadMesh at 0x7f3ce35de790>
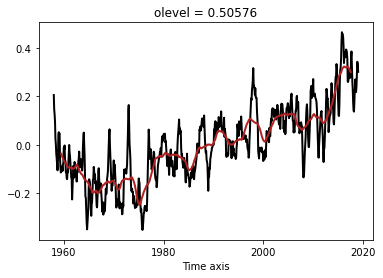
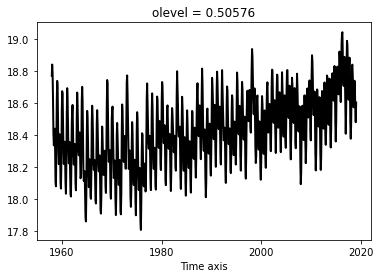
Compute the SST global mean time-series (weight by cell surface $e1t \times e2t$)
A first possibility would be to compute it using sum:
ts1 = (thetao * surface * tmask).sum(dim=['x', 'y']) / ((surface * tmask).sum(dim=['x', 'y']))
ts1.plot()
[<matplotlib.lines.Line2D at 0x7f3ce34d2ad0>]
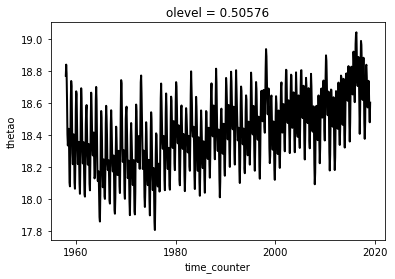
Another solution would be to use the xarray.weight method:
theta_weighted = thetao.weighted(surface * tmask)
ts2 = theta_weighted.mean(dim=['x', 'y'])
ts2.plot()
[<matplotlib.lines.Line2D at 0x7f3ce3460bd0>]
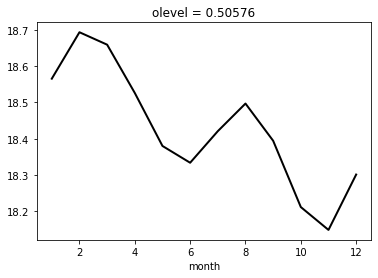
Remove the monthly clim from the time-series using
groupyontime_counter.month
clim = ts1.groupby('time_counter.month').mean(dim='time_counter')
clim.plot()
[<matplotlib.lines.Line2D at 0x7f3ce15c4d10>]
anom = ts1.groupby('time_counter.month') - clim
anom.plot()
[<matplotlib.lines.Line2D at 0x7f3ce1553350>]
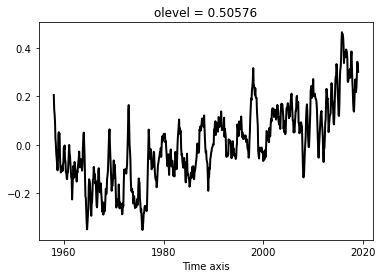
Compute the rolling mean of the time-series, using a 3-year window. Plot the raw and smoothed anomalies
tsroll = anom.rolling(time_counter=3*12 + 1, center=True).mean(dim='time_counter').dropna('time_counter')
tsroll
/home/barrier/Softwares/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:1: DeprecationWarning: Reductions are applied along the rolling dimension(s) '['time_counter']'. Passing the 'dim' kwarg to reduction operations has no effect.
"""Entry point for launching an IPython kernel.
<xarray.DataArray (time_counter: 696)>
array([-3.75917894e-02, -4.31766418e-02, -4.79905417e-02, -5.27248509e-02,
-5.73909217e-02, -6.10540841e-02, -6.47991929e-02, -6.75880931e-02,
-7.21680041e-02, -7.47213015e-02, -7.54183574e-02, -7.59560522e-02,
-7.97205163e-02, -8.48085868e-02, -8.79397065e-02, -9.16106189e-02,
-9.29940113e-02, -9.36864318e-02, -9.36281778e-02, -9.49310480e-02,
-9.54522487e-02, -9.61445246e-02, -9.57063573e-02, -9.53566924e-02,
-9.48523413e-02, -9.53026398e-02, -9.66476798e-02, -9.91032265e-02,
-9.92763432e-02, -9.90789530e-02, -9.77332073e-02, -9.69393097e-02,
-9.60886126e-02, -9.44236482e-02, -9.15832690e-02, -8.76608436e-02,
-8.42662787e-02, -8.31027624e-02, -8.42150040e-02, -8.67124815e-02,
-9.11820048e-02, -9.50429799e-02, -9.93276657e-02, -1.04367369e-01,
-1.10019382e-01, -1.12589072e-01, -1.15500044e-01, -1.19030303e-01,
-1.21328102e-01, -1.22604122e-01, -1.23015603e-01, -1.25352343e-01,
-1.28752738e-01, -1.32901609e-01, -1.37672675e-01, -1.41331329e-01,
-1.43540661e-01, -1.45733089e-01, -1.46563607e-01, -1.47840023e-01,
-1.48443398e-01, -1.49906927e-01, -1.52963849e-01, -1.54386801e-01,
-1.56194641e-01, -1.58370392e-01, -1.62440335e-01, -1.67565708e-01,
-1.72405804e-01, -1.75409051e-01, -1.77953418e-01, -1.80622847e-01,
-1.84090128e-01, -1.89183492e-01, -1.94397083e-01, -1.98576885e-01,
-1.99842797e-01, -1.98386848e-01, -1.97764606e-01, -1.96550245e-01,
...
1.13885794e-01, 1.13260639e-01, 1.12044677e-01, 1.11914591e-01,
1.11120051e-01, 1.10989005e-01, 1.10150800e-01, 1.07895851e-01,
1.01968726e-01, 9.86244181e-02, 9.58485868e-02, 9.35590346e-02,
9.12008384e-02, 8.94269236e-02, 9.01981249e-02, 9.20260853e-02,
9.38121638e-02, 9.49175852e-02, 9.73895309e-02, 1.00339364e-01,
1.03354717e-01, 1.07076241e-01, 1.10727832e-01, 1.15071020e-01,
1.20330536e-01, 1.25463041e-01, 1.29952811e-01, 1.35375177e-01,
1.40159156e-01, 1.44918815e-01, 1.48771425e-01, 1.52627357e-01,
1.56951593e-01, 1.62098744e-01, 1.67139967e-01, 1.74623977e-01,
1.82149147e-01, 1.88368806e-01, 1.95039813e-01, 2.01657860e-01,
2.07992544e-01, 2.14542722e-01, 2.21799687e-01, 2.28638689e-01,
2.35899318e-01, 2.43619466e-01, 2.51841529e-01, 2.59294592e-01,
2.65847732e-01, 2.72574016e-01, 2.79693848e-01, 2.84327181e-01,
2.87743694e-01, 2.90866446e-01, 2.92835605e-01, 2.95162731e-01,
2.98732161e-01, 3.04242696e-01, 3.10189504e-01, 3.15237350e-01,
3.17775065e-01, 3.18825196e-01, 3.21046825e-01, 3.23766615e-01,
3.23404495e-01, 3.21989999e-01, 3.21035832e-01, 3.20007661e-01,
3.19042599e-01, 3.19483519e-01, 3.21134471e-01, 3.22902532e-01,
3.23083392e-01, 3.21216715e-01, 3.18441935e-01, 3.15436327e-01,
3.11922124e-01, 3.08617943e-01, 3.05246494e-01, 3.01030139e-01])
Coordinates:
olevel float32 0.5058
time_centered (time_counter) object 1959-07-16 12:00:00 ... 2017-06-16 0...
* time_counter (time_counter) object 1959-07-16 12:00:00 ... 2017-06-16 0...
month (time_counter) int64 7 8 9 10 11 12 1 2 ... 11 12 1 2 3 4 5 6- time_counter: 696
- -0.03759 -0.04318 -0.04799 -0.05272 ... 0.3119 0.3086 0.3052 0.301
array([-3.75917894e-02, -4.31766418e-02, -4.79905417e-02, -5.27248509e-02, -5.73909217e-02, -6.10540841e-02, -6.47991929e-02, -6.75880931e-02, -7.21680041e-02, -7.47213015e-02, -7.54183574e-02, -7.59560522e-02, -7.97205163e-02, -8.48085868e-02, -8.79397065e-02, -9.16106189e-02, -9.29940113e-02, -9.36864318e-02, -9.36281778e-02, -9.49310480e-02, -9.54522487e-02, -9.61445246e-02, -9.57063573e-02, -9.53566924e-02, -9.48523413e-02, -9.53026398e-02, -9.66476798e-02, -9.91032265e-02, -9.92763432e-02, -9.90789530e-02, -9.77332073e-02, -9.69393097e-02, -9.60886126e-02, -9.44236482e-02, -9.15832690e-02, -8.76608436e-02, -8.42662787e-02, -8.31027624e-02, -8.42150040e-02, -8.67124815e-02, -9.11820048e-02, -9.50429799e-02, -9.93276657e-02, -1.04367369e-01, -1.10019382e-01, -1.12589072e-01, -1.15500044e-01, -1.19030303e-01, -1.21328102e-01, -1.22604122e-01, -1.23015603e-01, -1.25352343e-01, -1.28752738e-01, -1.32901609e-01, -1.37672675e-01, -1.41331329e-01, -1.43540661e-01, -1.45733089e-01, -1.46563607e-01, -1.47840023e-01, -1.48443398e-01, -1.49906927e-01, -1.52963849e-01, -1.54386801e-01, -1.56194641e-01, -1.58370392e-01, -1.62440335e-01, -1.67565708e-01, -1.72405804e-01, -1.75409051e-01, -1.77953418e-01, -1.80622847e-01, -1.84090128e-01, -1.89183492e-01, -1.94397083e-01, -1.98576885e-01, -1.99842797e-01, -1.98386848e-01, -1.97764606e-01, -1.96550245e-01, ... 1.13885794e-01, 1.13260639e-01, 1.12044677e-01, 1.11914591e-01, 1.11120051e-01, 1.10989005e-01, 1.10150800e-01, 1.07895851e-01, 1.01968726e-01, 9.86244181e-02, 9.58485868e-02, 9.35590346e-02, 9.12008384e-02, 8.94269236e-02, 9.01981249e-02, 9.20260853e-02, 9.38121638e-02, 9.49175852e-02, 9.73895309e-02, 1.00339364e-01, 1.03354717e-01, 1.07076241e-01, 1.10727832e-01, 1.15071020e-01, 1.20330536e-01, 1.25463041e-01, 1.29952811e-01, 1.35375177e-01, 1.40159156e-01, 1.44918815e-01, 1.48771425e-01, 1.52627357e-01, 1.56951593e-01, 1.62098744e-01, 1.67139967e-01, 1.74623977e-01, 1.82149147e-01, 1.88368806e-01, 1.95039813e-01, 2.01657860e-01, 2.07992544e-01, 2.14542722e-01, 2.21799687e-01, 2.28638689e-01, 2.35899318e-01, 2.43619466e-01, 2.51841529e-01, 2.59294592e-01, 2.65847732e-01, 2.72574016e-01, 2.79693848e-01, 2.84327181e-01, 2.87743694e-01, 2.90866446e-01, 2.92835605e-01, 2.95162731e-01, 2.98732161e-01, 3.04242696e-01, 3.10189504e-01, 3.15237350e-01, 3.17775065e-01, 3.18825196e-01, 3.21046825e-01, 3.23766615e-01, 3.23404495e-01, 3.21989999e-01, 3.21035832e-01, 3.20007661e-01, 3.19042599e-01, 3.19483519e-01, 3.21134471e-01, 3.22902532e-01, 3.23083392e-01, 3.21216715e-01, 3.18441935e-01, 3.15436327e-01, 3.11922124e-01, 3.08617943e-01, 3.05246494e-01, 3.01030139e-01]) - olevel()float320.5058
- name :
- olevel
- long_name :
- Vertical T levels
- units :
- m
- positive :
- down
- bounds :
- olevel_bounds
array(0.50576, dtype=float32)
- time_centered(time_counter)object1959-07-16 12:00:00 ... 2017-06-...
- standard_name :
- time
- long_name :
- Time axis
- time_origin :
- 1958-01-01 00:00:00
- bounds :
- time_centered_bounds
array([cftime.DatetimeNoLeap(1959, 7, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 8, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 12, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 3, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 5, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 7, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 8, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1960, 12, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1961, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1961, 2, 15, 0, 0, 0, 0, has_year_zero=True), ... cftime.DatetimeNoLeap(2015, 12, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 3, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 5, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 7, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 8, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2016, 12, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 3, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 5, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 6, 16, 0, 0, 0, 0, has_year_zero=True)], dtype=object) - time_counter(time_counter)object1959-07-16 12:00:00 ... 2017-06-...
- axis :
- T
- standard_name :
- time
- long_name :
- Time axis
- time_origin :
- 1958-01-01 00:00:00
- bounds :
- time_counter_bounds
array([cftime.DatetimeNoLeap(1959, 7, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 8, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 9, 16, 0, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(2017, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 5, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 6, 16, 0, 0, 0, 0, has_year_zero=True)], dtype=object) - month(time_counter)int647 8 9 10 11 12 1 ... 12 1 2 3 4 5 6
array([ 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, ... 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6])
anom.plot(label='raw')
tsroll.plot(label='smoothed')
[<matplotlib.lines.Line2D at 0x7f3ce14dad50>]