EOF analysis¶
EOF analysis is performed using the Eofs package. In the following, the steps for the computation of an EOF decomposition is provided. The objective would be to compute the El Nino index based on the SST of the Northern Pacific.
Extraction of Pacific mask¶
The Pacific mask can first be extracted based on coordinates (longitudes and latitudes) as follows:
import xarray as xr
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams['text.usetex'] = False
data = xr.open_dataset('data/mesh_mask_eORCA1_v2.2.nc')
data = data.isel(z=0, t=0)
lon = data['glamt'].values
lat = data['gphit'].values
mask = data['tmask'].values
# converts lon from Atl to Pac.
lon[lon < 0] += 360
# mask based on latitudes
ilat = (lat <= 60) & (lat >= -20)
mask[~ilat] = 0
# mask based on longitudes
ilon = (lon >= 117 ) & (lon <= 260)
mask[~ilon] = 0
# extracting the domain using the slices
ilat, ilon = np.nonzero(mask == 1)
ilat = slice(ilat.min(), ilat.max() + 1)
ilon = slice(ilon.min(), ilon.max() + 1)
mask = mask[ilat, ilon]
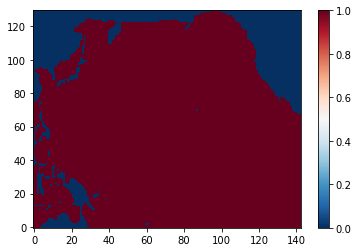
plt.figure()
cs = plt.imshow(mask, interpolation='none')
cb = plt.colorbar(cs)
Computation of seasonal anomalies¶
Now, we need to compute the seasonal anomalies of SST fields. First, we read the SST values and extract the spurious olevel dimension.
data = xr.open_dataset("data/surface_thetao.nc")
data = data.isel(olevel=0, x=ilon, y=ilat)
ntime = data.dims['time_counter']
data = data['thetao']
Now, we compute the anomalies using the groupy methods:
clim = data.groupby('time_counter.month').mean()
anoms = data.groupby('time_counter.month') - clim
Detrending the time-series¶
Now that the anomalies have been computed, the linear trend is removed using the detrend function. Since the detrend function does not manage NaNs, the filled values are first replaced by 0s
import scipy.signal as sig
import time
anoms = anoms.fillna(0)
anoms_detrend = sig.detrend(anoms, axis=0)
print(type(anoms_detrend))
<class 'numpy.ndarray'>
Note that in the detrend function returns a numpy.array object. Hence, no benefit will be taken from the xarray structure in the EOF calculation.
Extracting the weights¶
Now, the next step is to extract the weights for the EOFs, based on the cell surface and mask.
mesh = xr.open_dataset('data/mesh_mask_eORCA1_v2.2.nc')
mesh = mesh.isel(t=0, x=ilon, y=ilat)
surf = mesh['e1t'] * mesh['e2t']
surf = surf.data * mask # surf in Pacific, 0 elsewhere
weights = surf / np.sum(surf) # normalization of weights
Since EOF are based on covariance, the root-square of the weights must be used.
weights = np.sqrt(weights)
Computation of EOFS (standard mode)¶
The EOFS can now be computed. First, an EOF solver must be initialized. The time dimension must always be the first one when using numpy.array as inputs.
import eofs
from eofs.standard import Eof
solver = Eof(anoms_detrend, weights=weights)
Now, EOF components can be extracted. First, the covariance maps are extracted.
neofs = 2
nlat, nlon = surf.shape
covmaps = solver.eofsAsCovariance(neofs=neofs)
print(type(covmaps))
plt.figure()
plt.subplot(211)
cs = plt.imshow(covmaps[0], cmap=plt.cm.RdBu_r)
cs.set_clim(-1, 1)
cb = plt.colorbar(cs)
plt.subplot(212)
cs = plt.imshow(covmaps[1], cmap=plt.cm.RdBu_r)
cs.set_clim(-1, 1)
cb = plt.colorbar(cs)
<class 'numpy.ndarray'>
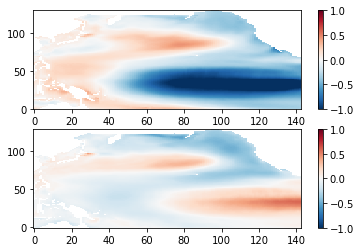
Then, we can recover the explained variance:
eofvar = solver.varianceFraction(neigs=neofs) * 100
eofvar
array([32.90275273, 7.66532732])
Finally, we can obtain the principal components. To obtain normalized time-series, the pscaling argument must be equal to 1.
pcs = solver.pcs(pcscaling=1, npcs=neofs).T
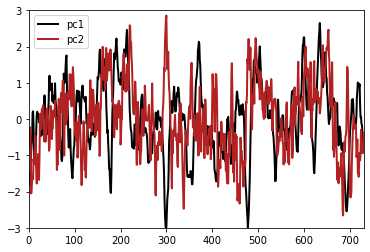
plt.figure()
plt.plot(pcs[0], label='pc1')
plt.plot(pcs[1], label='pc2')
leg = plt.legend()
plt.gca().set_ylim(-3, 3)
plt.gca().set_xlim(0, len(pcs[0]) - 1)
plt.savefig('ts1')
EOF computation (xarray mode)¶
In order to have EOF as an xarray with all its features, the Eof method of the eofs.xarray submodule must be used.
from eofs.xarray import Eof
Since it uses named labels, the time_counter dimension must first be renamed in time:
anoms = anoms.rename({'time_counter': 'time'})
anoms
<xarray.DataArray 'thetao' (time: 732, y: 130, x: 143)>
array([[[-0.31064034, -0.43762016, -0.6249218 , ..., -0.15113449,
-0.14532661, -0.12656021],
[-0.58628273, -0.527586 , -0.51573753, ..., -0.17450714,
-0.16910744, -0.16069221],
[-0.50172234, -0.5104065 , -0.45276833, ..., -0.22327042,
-0.2349453 , -0.24970245],
...,
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ]],
[[-0.01728249, -0.38983345, -0.48926353, ..., -0.6211891 ,
-0.5961971 , -0.57370186],
[ 0.22153282, 0.18938446, 0.04576683, ..., -0.6281853 ,
-0.6296387 , -0.6185684 ],
[ 0.43194962, 0.39792442, 0.34593582, ..., -0.63476753,
-0.68650436, -0.66425705],
...
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ]],
[[-1.4598808 , -1.3692341 , -1.0059109 , ..., 0.25130844,
0.290699 , 0.33873367],
[-1.3499966 , -1.3351994 , -1.1092415 , ..., 0.1412735 ,
0.18891907, 0.23643875],
[-1.2326813 , -1.1757793 , -1.0902805 , ..., 0.02304268,
0.05507469, 0.06399536],
...,
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ]]], dtype=float32)
Coordinates:
nav_lat (y, x) float32 -19.61 -19.61 -19.61 ... 55.74 55.78 55.82
nav_lon (y, x) float32 117.5 118.5 119.5 ... -103.1 -102.3 -101.4
olevel float32 0.5058
time_centered (time) object 1958-01-16 12:00:00 ... 2018-12-16 12:00:00
* time (time) object 1958-01-16 12:00:00 ... 2018-12-16 12:00:00
month (time) int64 1 2 3 4 5 6 7 8 9 10 ... 3 4 5 6 7 8 9 10 11 12
Dimensions without coordinates: y, x- time: 732
- y: 130
- x: 143
- -0.3106 -0.4376 -0.6249 -0.8991 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0 0.0
array([[[-0.31064034, -0.43762016, -0.6249218 , ..., -0.15113449, -0.14532661, -0.12656021], [-0.58628273, -0.527586 , -0.51573753, ..., -0.17450714, -0.16910744, -0.16069221], [-0.50172234, -0.5104065 , -0.45276833, ..., -0.22327042, -0.2349453 , -0.24970245], ..., [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[-0.01728249, -0.38983345, -0.48926353, ..., -0.6211891 , -0.5961971 , -0.57370186], [ 0.22153282, 0.18938446, 0.04576683, ..., -0.6281853 , -0.6296387 , -0.6185684 ], [ 0.43194962, 0.39792442, 0.34593582, ..., -0.63476753, -0.68650436, -0.66425705], ... [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[-1.4598808 , -1.3692341 , -1.0059109 , ..., 0.25130844, 0.290699 , 0.33873367], [-1.3499966 , -1.3351994 , -1.1092415 , ..., 0.1412735 , 0.18891907, 0.23643875], [-1.2326813 , -1.1757793 , -1.0902805 , ..., 0.02304268, 0.05507469, 0.06399536], ..., [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]]], dtype=float32) - nav_lat(y, x)float32-19.61 -19.61 ... 55.78 55.82
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- bounds :
- bounds_nav_lat
array([[-19.605793, -19.605793, -19.605793, ..., -19.605793, -19.605793, -19.605793], [-18.66211 , -18.66211 , -18.66211 , ..., -18.66211 , -18.66211 , -18.66211 ], [-17.715097, -17.715097, -17.715097, ..., -17.715097, -17.715097, -17.715097], ..., [ 55.07081 , 55.30693 , 55.541912, ..., 55.08865 , 55.120705, 55.158985], [ 55.461082, 55.70359 , 55.9449 , ..., 55.419174, 55.452374, 55.492016], [ 55.847755, 56.09659 , 56.344177, ..., 55.743164, 55.777523, 55.818542]], dtype=float32) - nav_lon(y, x)float32117.5 118.5 119.5 ... -102.3 -101.4
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- bounds :
- bounds_nav_lon
array([[ 117.5 , 118.5 , 119.5 , ..., -102.5 , -101.5 , -100.5 ], [ 117.5 , 118.5 , 119.5 , ..., -102.5 , -101.5 , -100.5 ], [ 117.5 , 118.5 , 119.5 , ..., -102.5 , -101.5 , -100.5 ], ..., [ 112.26882 , 113.18725 , 114.11117 , ..., -103.07745 , -102.20534 , -101.33294 ], [ 111.95293 , 112.8657 , 113.784294, ..., -103.11195 , -102.24751 , -101.382805], [ 111.62308 , 112.52994 , 113.44295 , ..., -103.147865, -102.291435, -101.43473 ]], dtype=float32) - olevel()float320.5058
- name :
- olevel
- long_name :
- Vertical T levels
- units :
- m
- positive :
- down
- bounds :
- olevel_bounds
array(0.50576, dtype=float32)
- time_centered(time)object1958-01-16 12:00:00 ... 2018-12-...
- standard_name :
- time
- long_name :
- Time axis
- time_origin :
- 1958-01-01 00:00:00
- bounds :
- time_centered_bounds
array([cftime.DatetimeNoLeap(1958, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 3, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 5, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 7, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 8, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 12, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 3, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 5, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 7, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1959, 8, 16, 12, 0, 0, 0, has_year_zero=True), ... cftime.DatetimeNoLeap(2017, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 7, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 8, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2017, 12, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 3, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 5, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 7, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 8, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 12, 16, 12, 0, 0, 0, has_year_zero=True)], dtype=object) - time(time)object1958-01-16 12:00:00 ... 2018-12-...
- axis :
- T
- standard_name :
- time
- long_name :
- Time axis
- time_origin :
- 1958-01-01 00:00:00
- bounds :
- time_counter_bounds
array([cftime.DatetimeNoLeap(1958, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 3, 16, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(2018, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 12, 16, 12, 0, 0, 0, has_year_zero=True)], dtype=object) - month(time)int641 2 3 4 5 6 7 ... 6 7 8 9 10 11 12
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, ... 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
To make sure it works, coordinate variables need to be removed.
anoms = anoms.drop_vars(['nav_lat', 'nav_lon', 'time_centered', 'month'])
anoms
<xarray.DataArray 'thetao' (time: 732, y: 130, x: 143)>
array([[[-0.31064034, -0.43762016, -0.6249218 , ..., -0.15113449,
-0.14532661, -0.12656021],
[-0.58628273, -0.527586 , -0.51573753, ..., -0.17450714,
-0.16910744, -0.16069221],
[-0.50172234, -0.5104065 , -0.45276833, ..., -0.22327042,
-0.2349453 , -0.24970245],
...,
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ]],
[[-0.01728249, -0.38983345, -0.48926353, ..., -0.6211891 ,
-0.5961971 , -0.57370186],
[ 0.22153282, 0.18938446, 0.04576683, ..., -0.6281853 ,
-0.6296387 , -0.6185684 ],
[ 0.43194962, 0.39792442, 0.34593582, ..., -0.63476753,
-0.68650436, -0.66425705],
...
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ]],
[[-1.4598808 , -1.3692341 , -1.0059109 , ..., 0.25130844,
0.290699 , 0.33873367],
[-1.3499966 , -1.3351994 , -1.1092415 , ..., 0.1412735 ,
0.18891907, 0.23643875],
[-1.2326813 , -1.1757793 , -1.0902805 , ..., 0.02304268,
0.05507469, 0.06399536],
...,
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[ 0. , 0. , 0. , ..., 0. ,
0. , 0. ]]], dtype=float32)
Coordinates:
olevel float32 0.5058
* time (time) object 1958-01-16 12:00:00 ... 2018-12-16 12:00:00
Dimensions without coordinates: y, x- time: 732
- y: 130
- x: 143
- -0.3106 -0.4376 -0.6249 -0.8991 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0 0.0
array([[[-0.31064034, -0.43762016, -0.6249218 , ..., -0.15113449, -0.14532661, -0.12656021], [-0.58628273, -0.527586 , -0.51573753, ..., -0.17450714, -0.16910744, -0.16069221], [-0.50172234, -0.5104065 , -0.45276833, ..., -0.22327042, -0.2349453 , -0.24970245], ..., [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[-0.01728249, -0.38983345, -0.48926353, ..., -0.6211891 , -0.5961971 , -0.57370186], [ 0.22153282, 0.18938446, 0.04576683, ..., -0.6281853 , -0.6296387 , -0.6185684 ], [ 0.43194962, 0.39792442, 0.34593582, ..., -0.63476753, -0.68650436, -0.66425705], ... [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]], [[-1.4598808 , -1.3692341 , -1.0059109 , ..., 0.25130844, 0.290699 , 0.33873367], [-1.3499966 , -1.3351994 , -1.1092415 , ..., 0.1412735 , 0.18891907, 0.23643875], [-1.2326813 , -1.1757793 , -1.0902805 , ..., 0.02304268, 0.05507469, 0.06399536], ..., [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ], [ 0. , 0. , 0. , ..., 0. , 0. , 0. ]]], dtype=float32) - olevel()float320.5058
- name :
- olevel
- long_name :
- Vertical T levels
- units :
- m
- positive :
- down
- bounds :
- olevel_bounds
array(0.50576, dtype=float32)
- time(time)object1958-01-16 12:00:00 ... 2018-12-...
- axis :
- T
- standard_name :
- time
- long_name :
- Time axis
- time_origin :
- 1958-01-01 00:00:00
- bounds :
- time_counter_bounds
array([cftime.DatetimeNoLeap(1958, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 3, 16, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(2018, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 12, 16, 12, 0, 0, 0, has_year_zero=True)], dtype=object)
solver = Eof(anoms, weights=weights)
neofs = 2
covmaps = solver.eofsAsCovariance(neofs=neofs)
covmaps
<xarray.DataArray 'eofs' (mode: 2, y: 130, x: 143)>
array([[[ 0.10379874, 0.10604983, 0.06451281, ..., -0.350463 ,
-0.3487487 , -0.3477348 ],
[ 0.04711282, 0.03726971, 0.01815447, ..., -0.36571452,
-0.36313176, -0.3593843 ],
[ 0.01782429, 0.01496945, 0.00708527, ..., -0.3726195 ,
-0.36914334, -0.36383352],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]],
[[-0.11388557, -0.12287213, -0.13214622, ..., 0.00338123,
0.00419892, 0.00579085],
[-0.11459833, -0.12700635, -0.13634968, ..., 0.01180524,
0.01197228, 0.01189382],
[-0.11706407, -0.12831454, -0.13485081, ..., 0.01716777,
0.01456021, 0.01296214],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]]], dtype=float32)
Coordinates:
* mode (mode) int64 0 1
* y (y) int64 0 1 2 3 4 5 6 7 8 ... 121 122 123 124 125 126 127 128 129
* x (x) int64 0 1 2 3 4 5 6 7 8 ... 134 135 136 137 138 139 140 141 142
Attributes:
long_name: covariance_between_pcs_and_thetao- mode: 2
- y: 130
- x: 143
- 0.1038 0.106 0.06451 0.04754 nan nan nan ... nan nan nan nan nan nan
array([[[ 0.10379874, 0.10604983, 0.06451281, ..., -0.350463 , -0.3487487 , -0.3477348 ], [ 0.04711282, 0.03726971, 0.01815447, ..., -0.36571452, -0.36313176, -0.3593843 ], [ 0.01782429, 0.01496945, 0.00708527, ..., -0.3726195 , -0.36914334, -0.36383352], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-0.11388557, -0.12287213, -0.13214622, ..., 0.00338123, 0.00419892, 0.00579085], [-0.11459833, -0.12700635, -0.13634968, ..., 0.01180524, 0.01197228, 0.01189382], [-0.11706407, -0.12831454, -0.13485081, ..., 0.01716777, 0.01456021, 0.01296214], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - mode(mode)int640 1
- long_name :
- eof_mode_number
array([0, 1])
- y(y)int640 1 2 3 4 5 ... 125 126 127 128 129
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129]) - x(x)int640 1 2 3 4 5 ... 138 139 140 141 142
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142])
- long_name :
- covariance_between_pcs_and_thetao
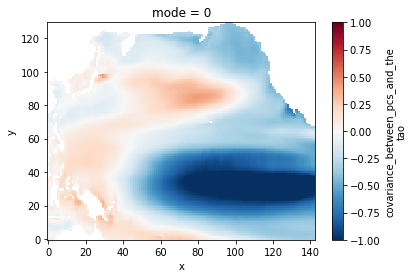
plt.figure()
cs = covmaps.isel(mode=0).plot()
cs.set_clim(-1, 1)
pcs = solver.pcs(pcscaling=1, npcs=neofs)
pcs
<xarray.DataArray 'pcs' (time: 732, mode: 2)>
array([[-1.0939478 , -0.23894596],
[-1.1724232 , -0.3829829 ],
[-1.382449 , -0.14822622],
...,
[-1.2102919 , -1.6217964 ],
[-1.3049158 , -1.6621873 ],
[-1.1621714 , -1.2294388 ]], dtype=float32)
Coordinates:
* time (time) object 1958-01-16 12:00:00 ... 2018-12-16 12:00:00
* mode (mode) int64 0 1- time: 732
- mode: 2
- -1.094 -0.2389 -1.172 -0.383 -1.382 ... -1.305 -1.662 -1.162 -1.229
array([[-1.0939478 , -0.23894596], [-1.1724232 , -0.3829829 ], [-1.382449 , -0.14822622], ..., [-1.2102919 , -1.6217964 ], [-1.3049158 , -1.6621873 ], [-1.1621714 , -1.2294388 ]], dtype=float32) - time(time)object1958-01-16 12:00:00 ... 2018-12-...
- axis :
- T
- standard_name :
- time
- long_name :
- Time axis
- time_origin :
- 1958-01-01 00:00:00
- bounds :
- time_counter_bounds
array([cftime.DatetimeNoLeap(1958, 1, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1958, 3, 16, 12, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(2018, 10, 16, 12, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2018, 12, 16, 12, 0, 0, 0, has_year_zero=True)], dtype=object) - mode(mode)int640 1
- long_name :
- eof_mode_number
array([0, 1])
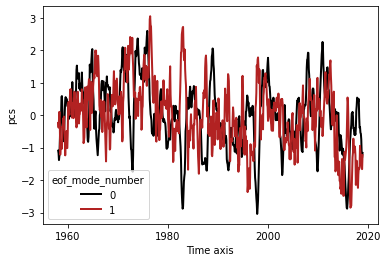
plt.figure()
l = pcs.plot.line(x='time')
plt.savefig('ts2')